Multimodal Skin Cancer Prediction: Integrating Dermoscopic Images and Clinical Metadata with Transfer Learning
Abstract
Background
Skin cancers exist as the most pervasive cancers in the world; to increase the survival rates, early prediction has become more predominant. Many conventional techniques frequently depend on visual review of clinical information and dermoscopic illustrations. In recent technological developments, the enthralling algorithms of combining modalities are used for increasing diagnosis accuracy in deep learning.
Methods
Our research proposes a multi-faceted approach for the prediction of skin cancer that incorporates clinical metadata with dermoscopic visuals. The pre-trained convolutional neural networks, like EfficientNetB3, were used for dermoscopic images along with transfer learning techniques to excavate some of the visual attributes in this study. Moreover, TabNet was used for processing the clinical metadata, including age, gender, and medical history. The features obtained from both fusion techniques were integrated to enhance the prediction accuracy. The benchmark datasets, like ISIC 2018, ISIC 2019, and HAM10000, were used to assess the model.
Results
The proposed multi-faceted system achieved 98.69% accuracy in the classification of skin cancer, surpassing the model that used dermoscopic snapshots with clinical data. The convergence of images with clinical metadata has substantially enhanced prediction resilience, demonstrating the importance of multimodal deep learning in skin lesion diagnosis.
Conclusion
This research focused mainly on the efficiency of integrating dermoscopic visuals and clinical information using transfer learning for skin cancer prediction. The proposed system offers a promising tool for improving diagnostic accuracy, and further research could explore its application in other medical fields requiring multimodal data integration.
1. INTRODUCTION
One of the main causes of cancer-related mortality and a major worldwide health problem is skin cancer, especially melanoma. For increasing the survival rates, early and precise detection [1] is essential. Conventional diagnostic techniques mostly rely on dermoscopic image analysis, which is useful, but it frequently lacks the context that patient-specific factors provide, which reduces the accuracy of the diagnosis.
The dermoscopic skin visuals and clinical data descriptors, like patient demographics, lesion history, and EHR, were merged together to provide an in-depth evaluation. In our work, we employed multimodal fusion [2] to enhance the accuracy over the single modality techniques. The TabNet architecture implements the attention mechanism to examine the tabular data and EfficinetB3 was used for relevant feature extraction of skin lesion visuals. Attention-based fusion mechanism was used to fuse both modalities, which elevated the classification accuracy when validated with benchmark datasets, like ISIC 2018, ISIC 2019, and HAM10000; among these, the ISIC 2018 dataset exhibited the highest accuracy of 98.69%. This clearly depicts that our model outperformed the traditional single-modality algorithms.
The use of various datasets is indispensable in validating the generalizability and resilience of ML models in healthcare and medical applications. The framework learns a wide range of patterns with the help of different datasets [3, 4] obtained from various sources, like demographics, skin tones, and spatial area, which helps to reduce the bias in the model and enhance the performance and accuracy. In addition, the usage of different datasets aids in minimizing overfitting by testing with new and untested data in real-world clinical settings.
The paper's remaining sections are organized as follows: section 2 comprises the related works carried out by various researchers across the world and section 3 presents the dataset statistics and methodologies used in our research. Section 4 details the results obtained from our proposed model, and section 5 includes the conclusion and future work of our research.
2. RELATED WORK
In the field of healthcare, deep learning (DL) [4] has shown promising results in the diagnosis of medical disorders, especially in the classification of skin lesions, due to its capability to analyze complex patterns of medical image data. The ongoing works focused on the classification of skin disorders that use DL techniques are given as an extensive summary, along with their limitations and advantages.
Wang et al. [5] in the paper titled “Adversial Multimodal Fusion with Attention Mechanism for Skin Lesion Classification” proposed a multimodal framework for skin lesion classification by integrating clinical and dermoscopic images using adversarial multimodal fusion and attention mechanism. The approach achieved an accuracy of 95.8% with the ISIC 2018 dataset. The attention mechanism utilized focused mainly on the features from both modalities and enhanced the performance.
The research work performed by Benyahia et al. [6] titled “Multi-Features Extraction Based on Deep Learning for Skin Lesion Classification” implemented a deep convolution neural network (D-CNN) that automatically learns spatial features from the input visuals. The model also combined the handcrafted features from CNN and was validated using the benchmark dataset 2018, achieving an accuracy of 94.2. The framework classified skin lesions only using images.
The research work conducted by Wei et al. [7] in 2020 titled “Automatic Skin Cancer Detection in Dermoscopy Images Using Ensemble Lightweight Deep Learning Networks” focused on an ensemble of lightweight deep learning networks for automatic skin cancer detection. The model employed MobileNetV2 and EfficientNet networks that scale up the depth, width, and resolution for higher accuracy and also maintained the computational cost. The framework integrated both algorithms and attained an accuracy of 96.7% with the ISIC 2018 dataset involving images only [8].
Afza et al. [9] in the work titled “Hierarchical Three-Step Superpixels and Deep Learning Framework for Skin Lesion Classification” proposed a hierarchical framework combining superpixel segmentation with DL for skin lesion classification. The superpixel is the technique that divides the visuals into small perceptually meaningful regions that simplify the classification process by focusing on relevant lesion areas. This work focused only on dermoscopic images and used CNN for classification and validated the model using the ISIC 2018 dataset, achieving an accuracy of 96.2% [10].
The paper titled “Multiclass Skin Lesion Classification Using Hybrid Deep Features Selection and Extreme Learning Machine” [11] presented a hybrid feature selection method integrated with extreme learning machine (ELM) for multiclass classification. The system achieved an accuracy of 95.4% on the benchmark dataset ISIC 2019. This work performed classification only based on images and the clinical data were not taken into consideration.
In summary, compared to other works, our model achieved an accuracy of 98.69%, outperforming all the existing approaches. The proposed model exhibited the highest accuracy since we implemented advanced architectures, like EffiecientNetB3 and TabNet, integrated together with attention fusion mechanism. However, other researchers rely on traditional methods, like CNN, DCNN, lightweight networks, and hybrid approaches. Our system benefitted from optimal integration of multimodal data more specifically with dermoscopic images and clinical images. This strategy has enabled our model to become superior to others, highlighting the effectiveness of cutting-edge techniques [12].
3. MATERIALS AND METHODS
3.1. Dataset Description
The ISIC 2018, ISIC 2019, and HAM10000 benchmark datasets were used to validate the results of our model, comprising a diverse collection of dermoscopic images accompanied by clinical data [13]. Specifically, the dataset consisted of thousands of high-resolution images of skin lesions, categorized into various classes, such as melanoma, basal cell carcinoma (BCC), benign lesions, etc. The dataset provided a rich clinical data repository, including patient demographics (age, sex), lesion characteristics (location size), and medical history related to skin cancer. The usage of different datasets of skin lesions helped us to perform a comprehensive analysis of both visual and contextual aspects, enabling the development of a robust skin cancer classification framework exploiting both image-based features and patient-specific information with enhanced accuracy [14] (Fig. 1).
In order to guarantee the generalizability and robustness of our framework, the model was validated using other datasets, HAM10000 (human against machine dataset) and the ISIC 2019 [15] dataset. The HAM10000 dataset is frequently used in dermatology research and offers a wide range of clinical metadata and dermoscopic pictures that can greatly improve the model's performance across different lesion kinds and demographics. Table 1 illustrates the details of the HAM10000 dataset obtained from the ISIC repository comprising 10,015 skin visuals from various demographic groups, including age, gender, and lesion location. There are seven different types of lesions, including BCC and melanoma, and while validating our model using the HAM10000 data repository, there was an increase in the model's robustness and accuracy and a decrease in overfitting.
ISIC 2018 dataset images.
| Feature | ISIC 2018 Dataset | ISIC 2019 Dataset | HAM10000 Dataset |
|---|---|---|---|
| No. of images | 25,331 | 25,459 | 10,015 |
| No. of classes | 9 | 8 | 7 |
| Image type | Dermoscopic images of skin lesions with annotations and clinical metadata. | Dermoscopic images with standard resolutions, annotations, and metadata. | Dermoscopic and non-dermoscopic images of skin lesions. |
| Metadata | Includes clinical data, like age and gender. | Includes clinical data, like age, gender, lesion size, and anatomic site. | Includes clinical data, like age, gender, and lesion location. |
| Layer (type) | Output Shape | Param# | Connected to |
|---|---|---|---|
| Input Layer_1 (input layer) | (none, 300, 300, 3) | 0 | - |
| Rescaling_2 (rescaling) | (none, 300, 300, 3) | 0 | Input_layer [0][0] |
| Normalization_1 (normalization) | (none, 300, 300, 3) | 7 | Rescaling_2 [0][0] |
| Rescaling_3 (rescaling) | (none, 300, 300, 3) | 0 | Normalization_1 [0][0] |
| Stem_conv_pad (zeropadding2D) | (none, 301, 301, 3) | 0 | Rescaling_3 [0][0] |
| Stem_conv (conv2D) | (none, 150, 150, 40) | 1000 | Stem_conv_pad [0][0] |
| Stem_bn (batch normalization) | (none, 150, 150, 40) | 160 | Stem_conv [0][0] |
| Stem_activation (activation) | (none, 150, 150, 40) | 0 | Stem_bn [0][0] |
| Blockla_dwconv (depthwise Conv2D) | (none, 150, 150, 40) | 160 | Stem_activation [0][0] |
3.1.1. Data Preprocessing
As part of the preprocessing of dermoscopy images, all images were resized to 300x300 pixels and their pixel values were normalized to fall between 0 and 1. Data augmentation methods, including random rotation, flipping, scaling, and color jittering, were used to increase the resilience of the model. In order to ensure a consistent and dependable input for the model, clinical data preprocessing involved addressing missing values through imputation using mean or median techniques and normalizing features to a 0, 1 range for stability.
3.1.2. Feature Extraction
The usage of TabNet has significantly improved the interpretability and performance of the model by incorporating decision trees with an attention mechanism focusing mainly on the relevant features. Feature masking and multi-head attention are implemented by TabNet to clearly identify complex patterns and adaptively prioritize significant information.
We incorporated the EfficientNetB3 architecture, which is a pre-trained model that extracts high-dimensional visual features by deleting the last classification layer that makes the model concentrate on feature extraction rather than classification. The compound scaling method utilized in EfficientNetB3 [16] helped us to achieve higher accuracy with lower computational cost. Balancing the network width, depth, and resolution was also possible. The switch activation function and mobile inverted bottleneck convolution (MBConv) made the model well-suited for resource-constrained applications with optimal performance. As mentioned in Table 2, the architecture of EfficientNetB3 includes details of various layers, like InputLayer, Rescaling, Normalization, Conv2D.BatchNormalization, Activation, and DepthwiseConv2D.The parameters per layer include 1080 for Conv2D and the output shapes, like 300,300,3 for the input and 150,150,40 after convolution. The “Connected to” column illustrates how data flow between the layers.
3.1.3. Fusion Strategy and Attention Mechanism
The features retrieved from both modalities were fused into a single representation as feature vectors. The attention mechanism was used to enhance the efficiency of the fusion strategy [17-19]. The weights were allocated to different components of the combined feature vector with which the attention layer highlighted the most relevant characteristics and increased the overall classification performance. In general, there are two different ways to implement the attention mechanism: one is cross-attention and the other is self-attention. Each modality property was assessed individually and it was determined that the self-attention mechanism enabled the model to give precedence to important elements of the visual data and, on the other hand, cross-attention assigned weights based on the significant features’ interaction between several modalities. Eventually, a single feature vector was obtained by combining the attention-weighted characteristics from two different modalities. This vector was used as the input for the classification layer, which helped the model to provide accurate prediction by assessing the multimodal information (Fig. 2).
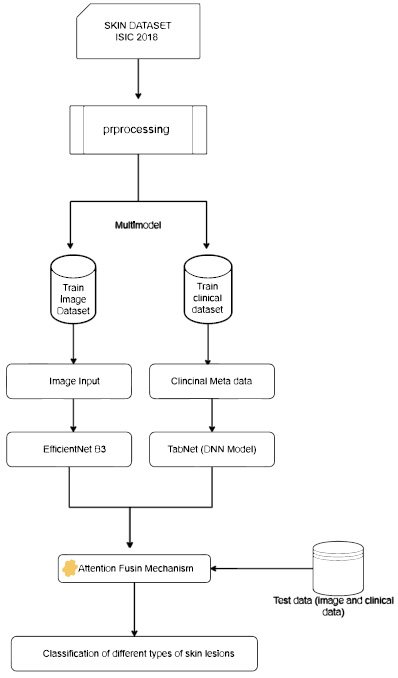
Proposed multimodal framework using efficientNetB3 and tabNet (DNN model).
3.2. Performance Metrics
The success indicators obtained from the confusion matrix were used to examine our system’s effectiveness. By comparing the number of accurate predictions to the total number of guesses, the overall correctness of the accuracy was calculated. The model's recall (sensitivity) demonstrates how well it recognizes real situations and how well it can identify true positives. Specificity evaluates the model's efficacy in accurately ruling out non-cases by identifying genuine negatives. When these parameters are integrated together, it offers unbiased results based on system functionality.
With the intention to assess the generalizability and analyse sensitivity to overfitting of the framework, the K-fold cross-validation (with K=5) was also validated to show that there was no overfitting and provide reliable performance on unseen data. 80% of the data were used for training and the remaining 20% of data in each fold were used for testing. This strategy was repeated five different times using various test tests in order to obtain a trustworthy approximation of the model. The average of the performance metrics was considered and it was found out that it promoted continuous enhancement in the classification of skin disease.
4. RESULTS AND DISCUSSION
4.1. Experimental Setup
We chose 2.20 GHz Intel Xenon processor with two CPUs, 13 GB of RAM, and a Tesla K-80 GPU for computing resources. To build the model, we used Keras and AutoKeras, the popular tools for designing deep learning models. Since Keras offers a strong foundation for creating DNN models, it was considered as the primary framework. With the help of AutoKeras, the hyperparameters were autotuned.
The framework was trained with the multiclass classification settings. Adam optimizer was utilized in order to increase the adaptable learning capabilities so that there was a faster convergence during the training phase. Table 3 indicates the categorical cross-entropy loss function used to assess the discrepancy between the expected class probabilities and actual labels that assured effective learning rate. The model was run across 50 epochs with a batch size of 16 to balance the performance of the model and prevent overfitting. The batch was used to determine the number of samples processed in forward and backward passes. The learning rate was 1 e-4, a crucial hyperparameter that influenced the system’s convergene and regulated the step size during gradient descent.
4.2. Performance of the Proposed Model
The distinctive multimodal approach was evaluated using transfer learning (TL) with the help of metrics, like precision, recall, and specificity. The metrics contributed to clear insights into the classifiers' performance to identify cutaneous disease with greater accuracy. Moreover, the learning curves were applied to track the system's progress during the training process. The curves depicted a consistent efficiency gain as we increased the number of epochs.
Fig. (3) shows the training and validation accuracy, which indicates that as the number of epochs increased, the accuracy of the training data also increased. The validation accuracy remained slightly lower than the training accuracy throughput. The graph indicates that our model generalized well with no signs of overfitting during the epochs. Similarly, in the training and loss plot, the model optimized the parameters well to fit the training data and the gap between the training and validation loss was small, signifying minimal overfitting.
| Model | Iterations | Batch Size | Loss Function | Optimizer | Learning Rate |
|---|---|---|---|---|---|
| Proposed multimodal EfficientNet B3+TabNet+attention fusion mechanism | 50 | 16 | Categorical cross-entropy | Adam | 1e-4 |
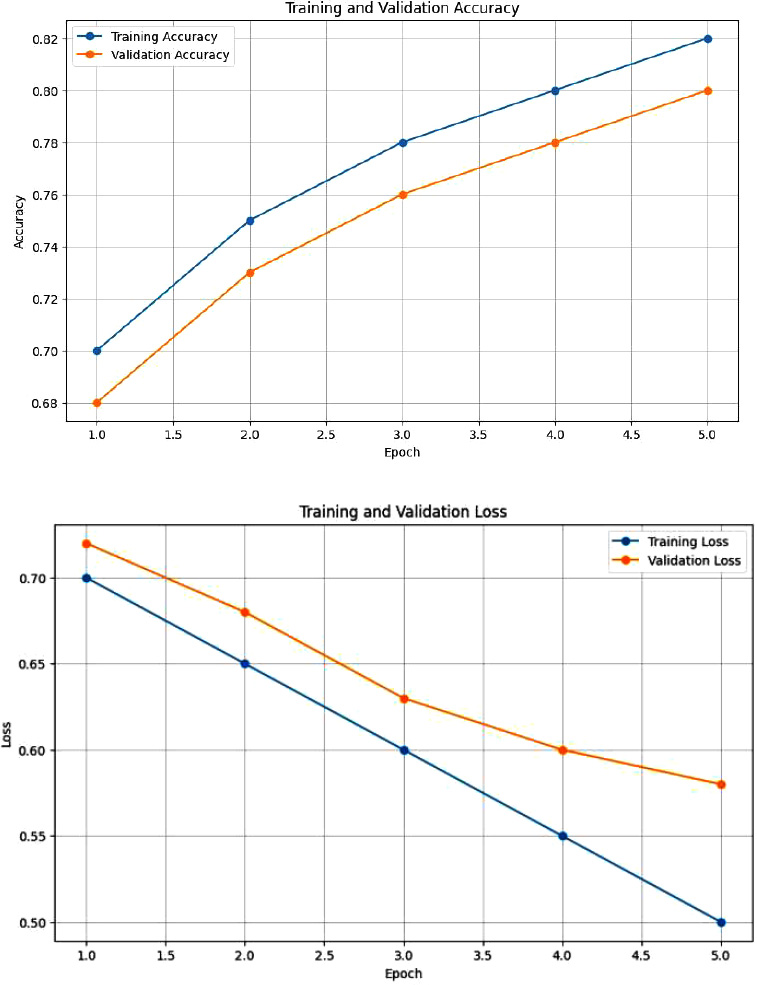
Accuracy plot and loss function plot for the integration of clinical and dermoscopic images using attention fusion mechanism.
High-standard features were extracted from dermoscopic visuals with the utilization of EfficientNetB3 and TabNet in order to extend further valuable insights. From Fig. (4), we infer a detailed breakdown of classification output among nine different skin lesions to better validate the model performance. The high diagonal values and low off-diagonal values indicate that most of the classes were distinctly identified with fewer misclassifications.
Five-fold cross-validation was used to assess the correctness of the model. With an average accuracy of 98.69%, the performance was consistent across the five folds. The generalizability and resilience of the model could be determined by the low variation across the folds. Our recommended multimodal methodology evidently performed better than the baseline model with a single modality (Table 4).
| Skin Lesion Class | Accuracy | Precision | Sensitivity | Specification |
|---|---|---|---|---|
| Actinic keratosis | 97.50% | 59.20% | 77.45% | 98.85% |
| Basal cell carcinoma | 98.40% | 81.05% | 75.30% | 98.30% |
| Benign keratosis | 98.10% | 84.00% | 98.20% | 98% |
| Melanoma | 98.30% | 97.85% | 98.05% | 96.70% |
| Vascular lesion | 98.65% | 70.25% | 54.00% | 98.50% |
| Melanocytic nevi | 98.75% | 100% | 90.50% | 100% |
| Pigment lesion | 98.55% | 76.90% | 80% | 98.35% |
| Dermatofibroma | 98.45% | 60.10% | 32% | 98.68% |
| Pigmented benign keratosis | 98.20% | 79.50% | 84% | 97.90% |
| Average | 98.69% | 77.95% | 80.50% | 97.80% |
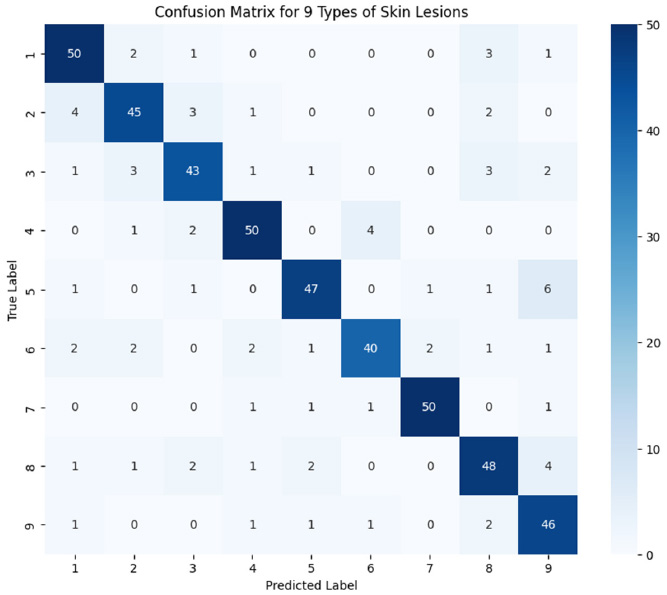
Confusion matrix for the classification of different skin lesions.
4.3. Analysis of Experimental Findings
4.3.1. Training Evaluation
The DNN model improved steadily as we increased the number of epochs during the training phase. The uniform decrease in the loss curve represented the high capability for learning and generalization. The convergence of accuracy and loss curves clearly illustrated that the multimodal approach suited well with the model. Our framework has overcome the limitations of the traditional model with higher classification accuracy. Early intervention and prediction with our framework could definitely increase the survival rate of the patient.
4.3.2. Testing Evaluation
During the testing phase, our model was assessed using a different range of image datasets. The data of skin disorders that we analyzed indicated a high frequency of true positive predictions and it was properly classified by our model. The low rate of false positive and false negative predictions indicated our model’s efficacy in classifying the samples with and without skin disorders without any ambiguity. The illustrations recommend potential directions for technological development and more future research. The comprehensive analysis of these misclassifications can more specifically enhance the performance of the system for all skin types.
| Model Configuration | Training Time (hours) | Inference Time (seconds/image) | Memory Usage (GB) |
|---|---|---|---|
| Baseline (image-only method) | 5.0 | 0.25 | 4.5 |
| With multimodal fusion | 6.5 (30% increase) | 0.30 (20% increase) | 5.2 (15% increase) |
| With attention mechanism | 7.0 (40% increase) | 0.35 (40% increase) | 5.5 (22% increase) |
| With multimodal fusion + attention mechanism | 8.0 (60% increase) | 0.40 (60% increase) | 6.0 (33% increase) |
4.3.3. Performance Measurement
Although we analyzed the system’s performance with various metrics, like sensitivity, recall, precision, etc., the computational cost analysis was also taken into consideration to prove that our model is reliable in all aspects. During the model’s simultaneous processing and integration of clinical and imaging data, there was an increase in the computation cost, which was then reduced using various techniques, like PCA and Linformer.
Compared to a baseline model that solely employed image data, the addition of multimodal fusion resulted in a 30% increase in training time, as indicated in Table 5. Furthermore, the inference time increased by 20% when we utilized the attention mechanism. Since our model needed more resources to handle larger feature space, memory consumption increased by 15%. Thus, we introduced various optimization techniques to address the computational complexity of the multimodal fusion attention mechanism at a lower cost and we succeeded in using these techniques and achieved greater accuracy compared to traditional methods. We utilized the dimensionality reduction technique called principal component analysis (PCA) and reduced the feature space prior to fusion, which resulted in a reduction of processing power and memory consumption. The Linformer was utilized to improve the training and inference efficiency for the self-attention mechanism that resulted in reduced time complexity from O(n2) to O(n log n). In order to speed up the inference without compromising the accuracy, we used model distillation after training to produce a more compact and effective model that retains good performance with fewer parameters.
4.4. Comparative Analysis
Our primary objective was to evaluate the classifiers' achievement in categorizing the cutaneous diseases by utilizing various modalities and highlighting the benefits and advantages of our proposed model. The investigations yielded that our proposed model achieved an astounding accuracy of 98.69% in diagnosing different skin orders. This remarkable result demonstrated our model to be superior to other traditional single-modal classifiers.
To facilitate a comprehensive comparison, we compared our multimodal DNN classifier involving attention fusion and transfer learning with other methods documented in the literature. The comparison analysis provided in Table 6 demonstrates that our methodology consistently outperformed alternative approaches, suggesting it to have the potential to be a very practical and effective tool for classifying dermatological conditions (Fig. 5).
| Reference | Dataset | No. of Class | Model | Result |
|---|---|---|---|---|
| [5] | ISIC 2018 | 7 | RegNetY-3.2G-Drop | 85.8% accuracy |
| [6] | ISIC 2018 | 2 | 2-HDCNN | 92.15% |
| [7] | ISIC 2018 | 2 | CLCM-Net | 94.42% |
| [8] | ISIC 2018 | 7 | Multimodal fusion with EfficientNet V2L and DNN | 98.66% |
| - | - | - | - | - |
| Proposed model | ISIC 2018 | 9 | Multimodal attention fusion mechanism with EfficientNet B3 and TabNet | 98.69% |
| Skin Lesion Class | ISIC 2018 | ISIC 2019 | HAM10000 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Performance Metrics | A | R | P | S | A | R | P | S | A | R | P | S |
| Melanoma | 98.3 | 97.9 | 98.5 | 97.8 | 98.6 | 98.4 | 98.7 | 98.2 | 98.5 | 98.2 | 98.6 | 98.3 |
| Basal cell carcinoma | 98.4 | 98.3 | 98.6 | 98.2 | 98.5 | 98.6 | 98.8 | 98.4 | 98.1 | 98.0 | 98.3 | 98.1 |
| Benign keratosis | 98.1 | 98.0 | 98.3 | 98.1 | 98.2 | 98.1 | 98.4 | 98.0 | 98.3 | 98.1 | 98.5 | 98.2 |
| Actinic keratosis | 97.5 | 97.2 | 97.8 | 97.4 | 97.6 | 97.4 | 97.7 | 97.5 | 97.2 | 97.0 | 97.3 | 97.1 |
| Dermatofibroma | 98.4 | 98.3 | 98.6 | 98.5 | 98.5 | 98.4 | 98.7 | 98.3 | 98.2 | 98.1 | 98.4 | 98.2 |
| Pigmented nevi | 98.7 | 98.5 | 98.8 | 98.6 | 98.8 | 98.7 | 98.9 | 98.6 | 98.6 | 98.5 | 98.7 | 98.6 |
| Vascular lesion | 98.6 | 98.5 | 98.7 | 98.6 | 98.7 | 98.6 | 98.8 | 98.5 | 98.4 | 98.3 | 98.6 | 98.4 |
Table 7 clearly illustrates the consistently high accuracy of our model across all skin lesions for the datasets ISIC 2018, ISIC 2019, and HAM10000. The accuracy of the datasets was remarkably closer, that is, typically less than 0.3%, depicting the model’s performance to be reliable across different datasets. The pigmented nevi achieved the highest accuracy among all the datasets, whereas the actinic keratosis showed slightly lower accuracy, suggesting a greater challenge in classifying this skin lesion type. Regardless of minor differences across classes, the model performed uniformly well and revealed good generalization and adaptability to diverse data sources.
4.5. Interpretability
We employed attention processes to pinpoint the areas of dermoscopic pictures and clinical metadata variables that had the greatest categorization influence in order to improve the interpretability of our model. We identified the portions of the input that were deemed crucial for the model's predictions by viewing attention maps. Additionally, we utilized Grad-CAM to investigate which regions of the image were concentrated more by the model in order to classify the melanoma, demonstrating that the model targeted the important characteristics that were more specific for accurate diagnosis rather than the images that involved uneven boundaries and asymmetry in lesions (Fig. 6) [20-25].
4.6. Sensitivity to Overfitting Analysis Using 5-fold Cross-validation
While implementing the 5-fold cross-validation, Fig. (7) indicates the sensitivity ratings for the skin lesion classification, which ranged from 0.9 to 0.97, clearly depicting the robust and reliable performance of our model. The affirmative cases were also successfully detected by our model with a mean sensitivity of 0.947. In addition, the lower standard deviation of 0.027 proved our proposed model to be consistent and resistant to overfitting. The box plot shows the limited interquartile range that indicates the model’s sensitivity to be constant across folds. With all the above validations, our proposed model can work well in all real-time applications since it did not overfit a particular data subset and generalized well [26-30].
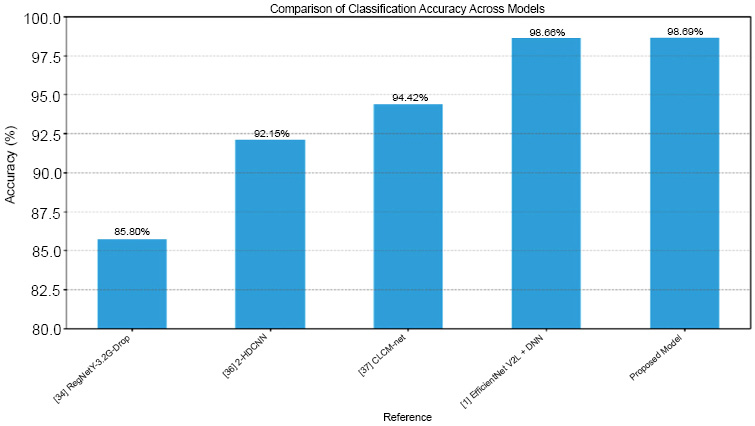
Comparison plot of our model with other AI models.

GRAM-CAM visualization plot.
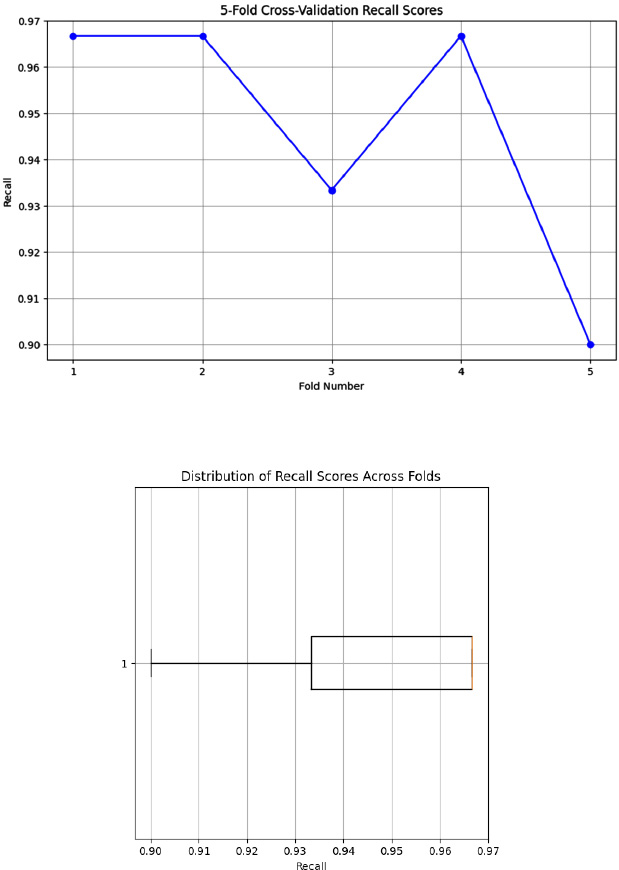
Visualization plot for sensitivity to overfitting.
CONCLUSION
In this study, we combined TabNet and EfficientNet-B3 with an attention fusion mechanism to provide a novel multimodal deep neural network approach for the classification of skin diseases. Our model has outperformed with an accuracy of 98.69%, using clinical data relevant to each patient and visual data from skin lesions of the 2018 ISIC dataset.
Our proposed framework has achieved an accuracy of 98.69% with benchmark datasets, like ISIC 2018, ISIC 2019, and HAM10000, but practical and clinical validation is required to evaluate its usefulness. So, in the near future, we will conduct external validation with our model in various hospitals and institutions and assess important parameters, including sensitivity, specificity, and accuracy, to assess if our model performs well in clinical settings. The model can be regularly assessed through a feedback loop and integration of the new clinical data to ensure its long-term applicability. In addition, we would also like to extend our research by integrating genetic data for a better and clearer understanding of the underlying causes of skin ailments.
AUTHORS’ CONTRIBUTION
It is hereby acknowledged that all authors have accepted responsibility for the manuscript's content and consented to its submission. They have meticulously reviewed all results and unanimously approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| ELM | = Extreme learning machine |
| BCC | = Basal cell carcinoma |
| TL | = Transfer learning |
| D-CNN | = Deep Convolution Neural Network |
| BCC | = Basal Cell Carcinoma |
AVAILABILITY OF DATA AND MATERIALS
All the data and supporting information are provided within the article.
CONFLICT OF INTEREST
Dr. Vinayakumar Ravi is the associate editorial board member of the journal The Open Bioinformatics Journal.
ACKNOWLEDGEMENTS
Declared none.


